Innowacje w diagnozie i leczeniu gruźlicy
Szukasz informacji o najnowszych metodach diagnozowania i leczenia gruźlicy? Znajdziesz je na predict-tb.eu

Różnorodność trumien i urn – co proponuje Dom Pogrzebowy Sokół?
Wybór odpowiedniej trumny lub urny stanowi kluczowy element organizacji ceremonii pogrzebowej. Dom pogrzebowy Sokół w Białymstoku oferuje szeroki asortyment, który zaspokaja różnorodne potrzeby oraz preferencje klientów. W artykule przedstawimy dostępne opcje, ich materiały oraz znaczenie symboliczn

Jak szybko i sprawnie przeprowadzić transakcję skupu telefonu?

Jakie innowacje technologiczne wpływają na rozwój wózków inwalidzkich?
Artykuły ostatnio dodane
Produkty i usługi dla każdego

Klamki okienne a ich wpływ na komfort użytkowania pomieszczeń
Wybór odpowiednich akcesoriów do okien ma kluczowe znaczenie dla komfortu użytkowania pomieszczeń. Różne modele klamek wpływają na funkcjonalność okien, co przekłada się na wygodę codziennego korzystania z przestrzeni. Przy wyborze warto zwrócić uwagę na estetykę, bezpieczeństwo oraz łatwość obsługi
Ochrona zdrowia i uroda
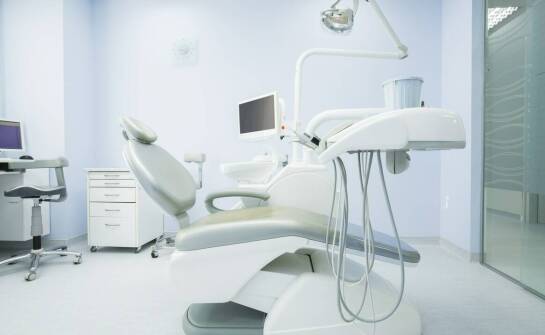
Jak wybrać odpowiedni stomatologiczny unit do swojego gabinetu?
Wybór odpowiedniego sprzętu stomatologicznego ma kluczowe znaczenie dla efektywności i komfortu pracy w gabinecie. Inwestycja w wysokiej jakości jednostki stomatologiczne przynosi liczne korzyści, zarówno dla lekarzy, jak i pacjentów. Nowoczesne technologie stosowane w tych urządzeniach zwiększają w
Transport, logistyka i pojazdy
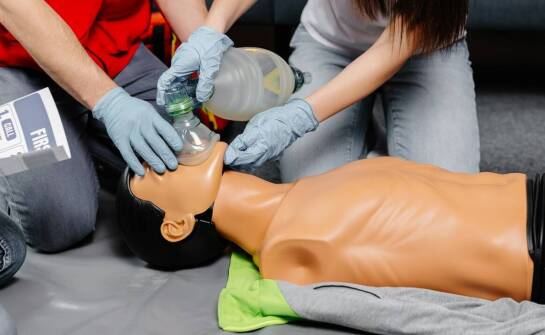
Kurs pierwszej pomocy w Płocku: nauka przez praktykę i doświadczenie
Kursy pierwszej pomocy w Płocku są kluczowe w sytuacjach kryzysowych. Uczestnicy uczą się, jak reagować w nagłych wypadkach dzięki doświadczonym instruktorom, co zwiększa ich pewność siebie oraz gotowość do działania. W artykule omówimy znaczenie praktyki w kursach pierwszej pomocy oraz korzyści pły
Zakupy, kultura i rozrywka

Przewodnik po akcesoriach do jadalni dostępnych w sklepie meblowym.
Akcesoria do jadalni odgrywają kluczową rolę w tworzeniu atmosfery oraz funkcjonalności tego pomieszczenia. Odpowiednie dodatki mogą podkreślić styl wnętrza i stworzyć przytulne miejsce do spożywania posiłków. W Galerii Wnętrz Reformy dostępny jest szeroki wybór produktów, które umożliwią aranżację
Elektronika, AGD, narzędzia

Jak montaż chłodni może wspierać rozwój zrównoważony firmy?
Montaż chłodni odgrywa kluczową rolę w zrównoważonym rozwoju, szczególnie w kontekście efektywności energetycznej oraz redukcji emisji CO2. Odpowiednie systemy chłodnicze znacząco obniżają zużycie energii, co przekłada się na niższe rachunki i mniejszy wpływ na środowisko. Firmy inwestujące w nowocz
Usługi i produkty remontowo-budowlane

Wpływ warunków eksploatacyjnych na nośność posadzki
Warunki eksploatacyjne odgrywają kluczową rolę w nośności posadzki, co ma znaczenie zarówno podczas projektowania, jak i użytkowania obiektów budowlanych. Przyjrzymy się czynnikom zewnętrznym, które mogą wpływać na trwałość oraz stabilność posadzek. Obciążenia, temperatura oraz wilgotność to element
Turystyka, noclegi i wypoczynek

Idealne miejsce na odpoczynek – hotel z basenem dla par
Wybór odpowiedniego miejsca na wypoczynek jest kluczowy dla par pragnących spędzić czas w romantycznej atmosferze. Hotel z basenem w Kujawsko-Pomorskiem stanowi idealną opcję, łącząc relaks z aktywnym spędzaniem czasu. Dzięki komfortowym pokojom, przestronnemu SPA oraz różnorodnym pakietom pobytowym
Odzież, obuwie i dodatki

Jakie materiały gwarantują trwałość roboczych spodni dla kobiet?
Wybór odpowiednich materiałów do produkcji odzieży roboczej jest kluczowy dla zapewnienia jej trwałości i funkcjonalności. W przypadku spodni roboczych damskich istotne jest, aby tkaniny spełniały wymagania różnych branż, takich jak produkcyjna, przemysłowa czy budowlana. Należy zwrócić uwagę na nor
Finanse i ubezpieczenia

Efektywne prowadzenie ryczałtu – usługi księgowego z Wrocławia
Efektywne prowadzenie ryczałtu jest kluczowe dla małych i średnich przedsiębiorstw, ponieważ wpływa na ich rozwój oraz stabilność finansową. Profesjonalna obsługa księgowa w Wrocławiu może znacząco ułatwić zarządzanie finansami, co przekłada się na oszczędność czasu i pieniędzy. Korzystanie z usług
Wnętrza - wyposażenie i wystrój

Ceramika bolesławiec w kontekście nowoczesnych wnętrz - jak ją wykorzystać?
Ceramika z Bolesławca to nie tylko tradycyjne wyroby, ale także element nowoczesnych wnętrz. Ręcznie zdobione produkty łączą estetykę z ekologicznymi właściwościami, co czyni je idealnym wyborem dla osób ceniących harmonię w aranżacji przestrzeni. W artykule omówimy różne sposoby wykorzystania ceram
Opieka i artykuły dziecięce

Jeździki na roczek a rozwój społeczny dziecka – jak zabawa wpływa na relacje?
Zabawa odgrywa niezwykle istotną rolę w rozwoju najmłodszych, a jeździki na roczek stanowią doskonały przykład aktywności wspierającej rozwój fizyczny i społeczny. W niniejszym artykule przyjrzymy się wpływowi tych zabawek na umiejętności społeczne dzieci oraz korzyści płynące z interakcji z rówieśn
Nauka i szkolnictwo

Jak terapeutyczna zabawa wpływa na emocjonalny rozwój dziecka?
Terapeutyczna zabawa jest nieoceniona w emocjonalnym rozwoju dzieci. Dzięki niej najmłodsi mogą wyrażać uczucia oraz uczyć się radzenia sobie z trudnymi emocjami. W artykule przyjrzymy się wpływowi zabawy na rozwój emocjonalny, a także skutecznym technikom terapeutycznym wykorzystywanym przez specja
Media i badanie rynku

Najnowsze trendy w projektowaniu ulotek i plakatów reklamowych
W dzisiejszym świecie reklama wizualna odgrywa kluczową rolę w przyciąganiu uwagi klientów. Ulotki i plakaty stanowią nie tylko nośniki informacji, ale także narzędzia do budowania wizerunku marki. Nowoczesne podejście do projektowania tych materiałów umożliwia skuteczniejszą komunikację z odbiorcam